Among 625 infants who were enrolled in a prospective 17-center cohort study—the 35th Multicenter Airway Research Collaboration (MARC-35) cohort—and had high-quality blood DNAm data measured at hospitalization before age 1 year, recurrent wheezing status before age 3 years was available for 488 infants, and asthma status by age 6 years was available for 506 infants (Fig. 1; Supplementary Fig. 1). These participants were included in the EWAS to identify differentially methylated regions (DMRs) associated with recurrent wheezing and asthma, respectively. Supplementary Data 1 shows the characteristics of these participants. Among participants with recurrent wheezing data, 170 (34.8%) developed recurrent wheezing by age 3 years; these infants were more likely to develop asthma by age 6 years compared to infants without recurrent wheezing (44.2% vs. 10.8%, p = 2.2 × 10−16). A total of 112 (22.1%) participants developed asthma by age 6 years in our study sample. We observed notable differences in several baseline variables between recurrent wheezing/asthma cases and non-cases. These variables include age at hospitalization, race/ethnicity, number of previous breathing problems, history of eczema, rhinovirus infection, and maternal asthma.
This study investigates the associations between epigenome-wide DNA methylation (794,125 high-quality CpG sites), measured using the Illumina Infinium MethylationEPIC array, in blood samples from infants with severe bronchiolitis, and the subsequent risk of recurrent wheezing (before 3 years of age, n = 448, 170 cases) and asthma (at 6 years of age, n = 506, 112 cases) during childhood. The analytical data were collected from participants of the 35th Multicenter Airway Research Collaboration (MARC-35) study. Thirty-one differentially methylated regions (DMRs) were associated with recurrent wheezing, and thirty-three DMRs were associated with asthma, taking into account potential interactions with rhinovirus infection during infancy. In silico blood cell-type deconvolution and chromatin state enrichment analyses were conducted to identify specific blood cell types likely driving the associations at these DMRs. The chromatin state annotation was obtained from the Roadmap Epigenomics Consortium. We then explored the relationship between these DMRs and the levels of 347 proteins measured in the same bronchiolitis infant blood samples using the Olink multiplex platform. For proteins associated with the DMRs, we constructed a protein-protein interaction (PPI) network leveraging data from the STRING database, performed two-sample mendelian randomization analysis using protein quantitative trait loci (pQTLs, UK Biobank and deCODE) and GWAS summary statistics from published studies, and conducted pathway enrichment analysis using data from the Gene Ontology database. These analyses aim to provide additional biological insights into the early development of childhood respiratory diseases in infants with severe bronchiolitis underlying the DNA methylation differences.
Epigenome-wide analysis identified novel DMRs associated with recurrent wheezing
We first examined the associations between DNAm and recurrent wheezing at 794,125 CpG sites that passed the quality control (Supplementary Fig. 2). No substantial genomic inflation was observed as indicated by the quantile-quantile plots (e.g., λgenomic control = 1.03 for linear regression, Supplementary Fig. 3). We identified 31 DMRs associated with recurrent wheezing after accounting for multiple testing (Šidák p < 0.01; Table 1, Supplementary Data 2, Supplementary Data 3). Of these, 21 DMRs were identified through the analysis based on linear regression, and additional 10 DMRs were identified through the analysis based on LRT-2df that leveraged potential interaction with rhinovirus infection to increase power. Ten DMRs exhibited different association patterns between participants who were rhinovirus-infected and those who were not (i.e., the nominal p-value < 0.05 for the recurrent wheezing×rhinovirus infection interaction at more than half of the CpG sites within a DMR).
The most significant recurrent wheezing DMR (chr18:13611190-13611807, Šidák p = 1.08 × 10−10; Table 1) was annotated to the LDLRAD4. Across all 11 CpG sites in this region, we observed a lower DNAm level among the recurrent wheezing cases compared with the non-cases, and the effect sizes were more pronounced among participants who had rhinovirus infection during infancy (i.e., 7 out of 11 CpG site in the DMR had nominal p-value < 0.05 for the interaction term).
Epigenome-wide analysis identified novel DMRs associated with childhood asthma
We then examined the associations between DNAm at 794,125 CpG sites and childhood asthma. No substantial genomic inflation was observed as indicated by the quantile-quantile plots (e.g., λgenomic control = 1.05 for linear regression, Supplementary Fig. 4). We identified 33 DMRs associated with asthma after accounting for multiple testing (Šidák p < 0.01; Table 2, Supplementary Data 4, Supplementary Data 5). Of these, 20 DMRs were identified through the analysis based on linear regression, and an additional 13 DMRs were identified through the analysis based on LRT-2df. Seven DMRs exhibited different association patterns between participants who were rhinovirus-infected and those who were not (i.e., the nominal p-value < 0.05 for the asthma×rhinovirus infection interaction at more than half of the CpG sites within a DMR).
The most significant asthma DMR (chr17:6899085-6899758, Šidák p = 1.31 × 10−13; Table 2) overlapped the promoter region of ALOX12. We observed a higher DNAm level among the asthma cases compared to the non-cases across all 12 CpG sites in this region. This asthma DMR overlapped with a recurrent wheezing DMR near ALOX12 (chr17:6898821-6899577, Šidák p = 3.15 × 10−8; Table 1). In addition, two other asthma DMRs were also associated with recurrent wheezing, with consistent direction of effects: chr6:28129313-28129656 (ZKSCAN8P1; higher DNAm in cases) and chr6:42927940-42928200 (GNMT; lower DNAm in cases).
Cell-type specific associations at the infant blood DMRs
The proportions of seven blood cell types (B cells, NK cells, CD4 T cells, CD8 T cells, monocytes, neutrophils, and eosinophils) were inferred based on the epigenome-wide DNAm data. Utilizing the CellDMC method15, which tests the interactions between the cell type fractions in each sample and the outcome of interest, we identified blood cell types that may drive the observed associations between the DMRs and the childhood respiratory outcomes.
Supplementary Fig. 5 illustrates the cell-type specific associations of DNAm with recurrent wheezing at selected DMRs (DMRs displaying nominal statistical significance, i.e., p < 0.05, for cell-type specific associations are shown). At the most statistically significant DMR for recurrent wheezing (chr18:13611190-13611807; LDLRAD4), the association appeared to be driven by effects in eosinophils, monocytes, and neutrophils. Other recurrent wheezing DMRs, such as chr2:54086854-54087343 (GPR75, ASB3; B cells), chr8:22132926-22133076 (PIWIL2; B cells), chr11:124622096-124622444 (VSIG2; neutrophils, eosinophils), and chr17:46681111-46681401 (HOXB6; CD4 T cells, CD8 T cells, eosinophils), also appeared to be influenced by cell-type-specific DNAm effects.
Supplementary Fig. 6 illustrates the cell-type specific associations of DNAm with asthma at selected DMRs with nominal statistical significance (p < 0.05) for cell-type specific associations. Asthma DMRs with evidence for cell-type specific DNAm effects include chr6:28058724-28058973 (ZSCAN12P1; CD4 T cells), chr7:95025855-95026248 (PON3; NK cells), chr16:66400320-66400599 (CDH5; monocytes, neutrophils, eosinophils), and chr16:67233432-67233983 (ELMO3; CD4 T cells).
DMRs for childhood respiratory outcomes were enriched in neutrophil-specific chromatin states
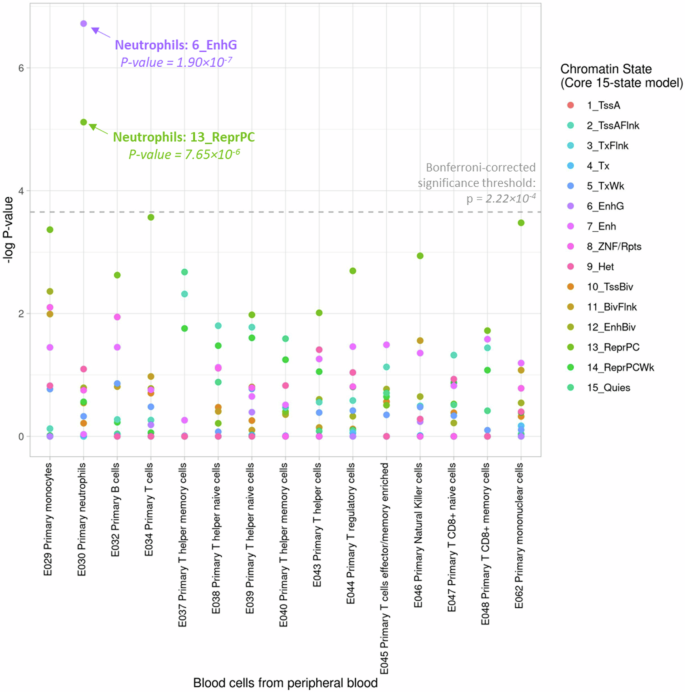
We conducted enrichment analysis to examine whether the 61 DMRs for recurrent wheezing and/or asthma were enriched with specific chromatin states in human peripheral blood cells from the Roadmap Epigenomics Project16. Our analysis (Fig. 2, Supplementary Data 6) revealed statistically significant enrichment of these DMRs in the Genic Enhancer (6_EnhG, p = 1.90 × 10−7) and Repressed Polycomb (13_ReprPC, p = 7.65 × 10−6) regions in primary neutrophils (Accession: E30), highlighting the potential roles of epigenetic regulation in neutrophils during early asthma development.
The 15 chromatin states were consolidated from the NIH Roadmap Epigenomics project based on five histone modification marks (H3K4me3, H3K4me1, H3K36me3, H3K9me3, H3K27me3) in human cells. Chromatin states are 1_TssA (Active TSS), 2_TssAFlnk (Flanking Active TSS), 3_TxFlnk (Transcr. at gene 5’ and 3’), 4_Tx (Strong transcription), 5_TxWk (Weak transcription), 6_EnhG (Genic enhancers), 7_Enh (Enhancers), 8_ZNF/Rpts (ZNF gene & repeats), 9_Het (Heterochromatin), 10_TssBiv (Bivalent/Poised TSS), 11_BivFlnk (Flanking Bivalent TSS/Enh), 12_EnhBiv (Bivalent Enhancer), 13_ReprPC (Repressed PolyComb), 14_ReprPCWk (Weak Repressed PolyComb), 15_Quies (Quiescent/Low). The enrichment p-value was obtained based on two-sided binomial test in 1000 permuted samples. To address multiple testing, we employed a Bonferroni-corrected significant threshold of p = 2.22 × 10−4 (the horizontal dashed line).
DMRs for childhood respiratory outcomes correlated with circulating levels of immune proteins in infant blood
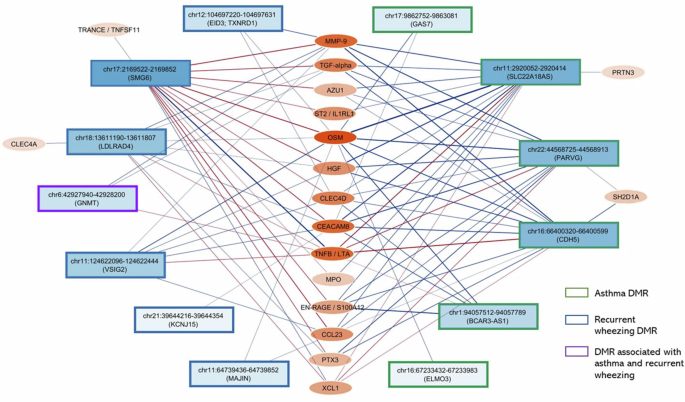
We examined the correlations between DNAm level at the DMRs and 347 immune-related proteins in infant blood, controlling for age, sex, race/ethnicity, and other key covariates. A total of 38 DMRs showed correlation with at least one protein level in infant blood (FDR < 0.05; Supplementary Data 1). Among them, the 4 DMRs correlated with the greatest number of proteins, namely, chr17:2169272-2169852 (SMG6, 51 proteins), chr22:44568725-44568913 (PARVG, 42 proteins), chr16:66400320-66400599 (CDH5, 41 proteins), and chr11:2920052-2920414 (SLC22A18AS, 37 proteins), shared correlation with the same set of 30 proteins (Supplementary Fig. 7a). These 30 proteins were also correlated with a large number of other DMRs. Some examples of these proteins are CEACAM8 (20 DMRs), MMP-9 (19 DMRs), TNFB (17 DMRs), AZU1 (17 DMRs), OSM (17 DMRs), HGF (17 DMRs), TGF-alpha (15 DMRs), and CLEC4D (15 DMRs) (Supplementary Fig. 7b).
To visually represent the DMR-protein correlations in infant blood, we created Fig. 3 to highlight the 18 proteins with the most statistically significant correlations with DMRs (FDR < 0.0005).
Rectangular nodes are DMRs, with the color shade corresponding to number of proteins the DMRs associated with. Oval nodes are proteins, with the color shade corresponding to number of DMRs the protein associated with. Edges represent associations between DMRs and proteins with a Benjamini-Hochberg FDR below 0.0005 after adjusting for covariates. The thickness of the edges corresponds to the statistical significance of the association (obtained from linear regression, two-sided test) calculated as average -log10(P) of the DNAm-protein correlation across all CpG sites within a DMR. Red edges show positive association. Blue edges show negative association.
Phenotypic associations between DMR-correlated proteins and asthma
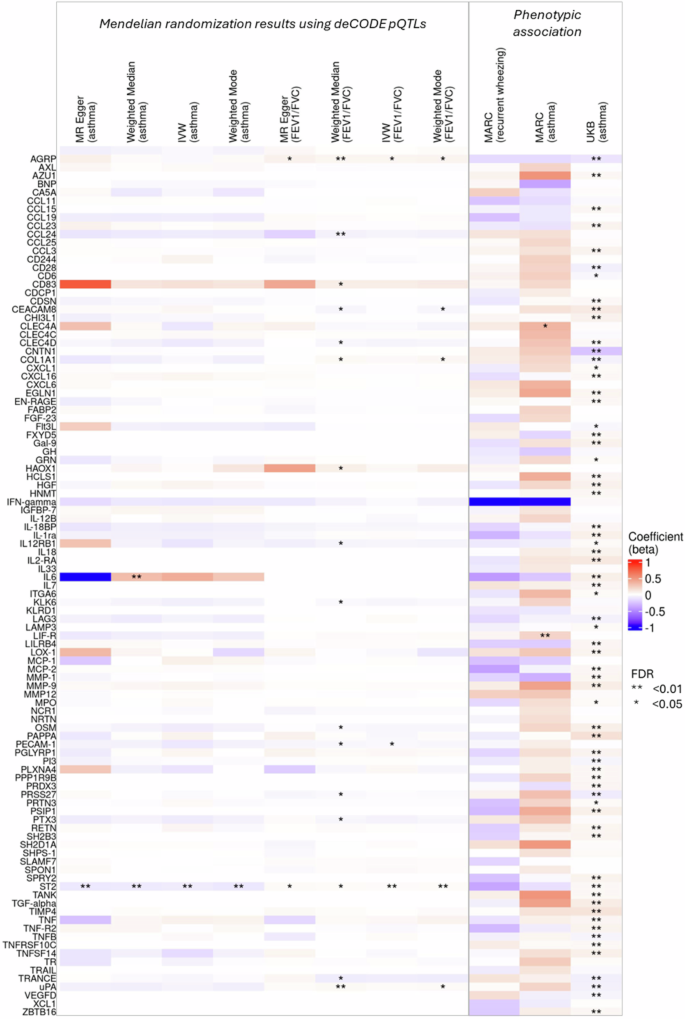
A total of 104 protein markers were statistically significantly (FDR < 0.05) correlated with the DMRs for childhood respiratory outcomes. Among these DMR-correlated proteins, levels of LIF-R and CHI3L1 in infant blood were prospectively associated with childhood asthma in a small subset of MARC-35 participants with available data (50 asthma cases, 59 non-cases) after correcting for multiple testing (FDR < 0.05; Fig. 4). Supplementary Data 8 provides detailed results for the phenotypic associations between protein levels and the respiratory outcomes. Using this data, we also found 6 and 24 proteins associated with later recurrent wheezing and asthma outcomes, respectively, with nominal significance (p < 0.05). Of them, IFN-γ was nominally associated with both recurrent wheezing (p = 0.006) and asthma (p = 0.032). We further leveraged recent findings from the UK Biobank (UKB)17 where associations between plasma proteomics and prevalent asthma were assessed in a larger population (N = 46,595; mean age: 57 years). Among all DMR-correlated proteins we identified, 63.5% (n = 66) also showed were associated with prevalent asthma in UKB (FDR < 0.05; Fig. 4, Supplementary Data 8).
Evidence for the putative causal relationship was obtained using two-sample mendelian randomization (MR Egger, weighted median, inverse variance weighted (IVW), weighted mode) with pQTLs from deCODE as genetic instruments. Phenotypic associations were obtained using linear regression. The analyses to identify phenotypic association in the MARC-35 cohort were conducted in a small subset of participants with available data (recurrent wheezing: 57 cases, 50 non-cases; asthma: 50 cases, 59 non-cases).
Protein–protein interactions and pathway enrichments of the DMR-correlated proteins
We derived protein-protein interaction (PPI) network from the STRING database18 based on previous experimental evidence and co-expression data (Supplementary Fig. 8). This network suggested that many DMR-correlated proteins may interact with each other. For instance, CEACAM8, AZU1, PRTN3, and MPO were significantly associated with DNAm at the DMRs (FDR < 0.0005). These proteins were also interconnected with each other through co-expression evidence, suggesting shared biological functions. Similarly, MMP-9, OSM, and HGF were interconnected with each other and also demonstrated evidence for interacting with other cytokines (e.g., IL-6, IL12RB1, IL18, TNF) and chemokines (e.g., CCL3, MCP-1/CCL2) through the PPI network. Additionally, we identified several pairs of known ligand-receptor interactions among the DMR-correlated proteins, including IL-18 and IL-18BP, IL-33 and ST2, IL-12B and IL12RB1, TNF and TNF-R2/TNFRSF1B, and TRAIL/TNFSF10 and TNFSFR10C.
The Gene Ontology (GO) pathway analyses for the 104 DMR-correlated proteins revealed multiple significantly enriched biological and molecular pathways (FDR < 0.05; Supplementary Data 9). Among the most significantly enriched pathways, several were related to the migration, differentiation, and proliferation of leukocytes or T cells. Other top enriched pathways included positive regulation of cell-cell adhesion (FDR = 7.36 × 10−16), cellular response to interferon-gamma (FDR = 2.55 × 10−12), cellular response to interleukin-1 (FDR = 9.73 × 10−11), regulation of tumor necrosis factor production (FDR = 8.22 × 10−6), and positive regulation of NIK/NF-κB signaling (FDR = 1.95 × 10−5).
Mendelian randomization for the DMR-correlated proteins and asthma-related outcomes
Utilizing protein quantitative trait loci (pQTLs) mapped in a recent study in the Icelandic deCODE population19, we conducted a two-sample mendelian randomization (MR) analysis to examine the causal relationship between the DMR-correlated proteins and asthma and lung function (FEV1/FVC) (Fig. 4, Supplementary Data 1,0). Notably, we found evidence supporting putative causal effects (FDR < 0.05) of plasma ST2 level on both asthma and FEV1/FVC through all MR methods we applied. Our results revealed that a higher plasma ST2 level may decrease the risk of asthma and increase lung function. These results remained robust in the sensitivity analyses when pQTLs derived from a recent UKB study17 were used as the genetic instruments (Supplementary Fig. 9, Supplementary Data 1,0), as well as when an alternative asthma genome-wide association study (GWAS) summary statistics were used in the MR analyses (Supplementary Data 11).
We further investigated whether ST2 may have causal effects on other asthma-related outcomes through MR. We found evidence supporting causal relationships between plasma ST2 level and allergic conditions, including allergic rhinitis and atopic dermatitis (Supplementary Data 11).
Additionally, our MR analyses suggested potential causal effects of plasma AGRP and CHI3L1 levels on FEV1/FVC when the deCODE pQTLs were used as genetic instruments; however, we did not find same results in the analysis using the UKB pQTLs (Supplementary Data 10).
Associations of DNAm with ST2 protein in infant blood
Given the consistent MR evidence suggesting potential causal effects of plasma ST2 protein level on asthma and related outcomes, we further evaluated the association of DNAm at the ST2 (also known as IL1RL1) locus with ST2 protein level in infant blood (n = 112, Supplementary Data 12). Among 19 CpG sites at this locus included on the EPIC array, DNAm at 6 CpG sites were significantly associated ST2 protein level after adjusting for key covariates (FDR < 0.05). When further adjusting for estimated blood cell types, the positive association between DNAm at cg17738684 (at distal promoter of ST2) and the ST2 protein remained statistically significant (FDR = 3.06 × 10−3). Here we note that DNAm at this CpG site was not associated with childhood asthma or recurrent wheezing outcomes in our study sample (Supplementary Data 12).
Additionally, at 8 DMRs associated with recurrent wheezing and/or asthma, we observed correlations between DNAm and the ST2 protein level in infant blood (FDR < 0.05, Supplementary Data 7). These DMRs are chr1:94057512-94057789 (BCAR3-AS1), chr11:2920052-2920414 (SLC22A18AS), chr11:124622096-124622444 (VSIG2), chr16:66400320-66400599 (CDH5), chr17:2169522-2169852 (SMG6), chr17:6899085-6899758 (ALOX12), chr18:13611190-13611807 (LDLRAD4), and chr22:44568725-44568913 (PARVG).