Sequence analysis of Spike protein from all SARS-CoV-2 variants
We have obtained the amino acid sequences of the spike protein from the original Wuhan_hCoV-19 strain and seven major SARS-CoV-2 variants from the protein database maintained by the National Center for Biotechnology Information (NCBI). Utilizing Clustal Omega, a multiple sequence alignment (MSA) was conducted to identify non-conserved regions. The results of the MSA are provided in Supplementary Fig. 1. This MSA file enabled the selection of predicted B-cell and T-cell epitopes within the conserved region among all SARS-CoV-2 variants.
Prediction of linear B-Cell epitopes
We employed the IEDB Analysis Resource to anticipate B-Cell epitopes through the evaluation of various parameters. Furthermore, we made use of the ABCpred and Bpred servers for the same purpose. Subsequently, we curated the epitopes by consolidating predictions from multiple servers to guarantee precision (Supplementary Table 1). Following a comprehensive analysis, we successfully determined 18 linear B-cell epitopes that were universally predicted by all the tools. Please refer to Table 1 for the detailed list.
Discontinuous epitope prediction
We employed DiscoTope, Ellipro, and SEPPA prediction tools to detect discontinuous epitopes in the SARS-CoV-2 S-protein. The Discotope score, based on the predictive capability of the algorithm, revealed a positive prediction for values exceeding − 3.7. The Ellipro method evaluates the 3D shape of proteins as a series of ellipsoids and computes a Protrusion Index (PI) score for each ellipsoid, identifying the highest scoring ellipsoid as the region of maximum surface accessibility. The SEPPA 3.0 prediction tool analyzes the 3D structure of the protein and assigns scores to individual residues based on the likelihood of being part of the epitope. Through these tools, we discovered potential epitope residues and predicted six discontinuous epitopes. Detailed information is available as supplementary data (Supplementary Fig. 2, Tables 2, 3 and 4). A consensus approach was employed to select common residues from these predictions. The most promising B-cell epitopes labelled, as B-Ep1 to B-Ep5 were designated based on their PI scores, indicating significant promise, especially for B-Ep1 to B-Ep4. Supplementary Fig. 3 illustrates the predicted discontinuous epitopes within the SARS-CoV2 S-protein structure (PDB: 6xe1).
Upon comparison of the anticipated linear and discontinuous epitopes, it was found that 53 residues from Table 2 are present in both prediction methods, implying a high likelihood of being part of the B-cell epitope. Except B-Ep2 and B-Ep3 residues, these epitopes were conserved in the spike protein among SARS-CoV-2 variants and are located closely to each other as depicted in Supplementary Fig. 3. This region plays a significant role in eliciting an immune response by binding to antibodies. The identified properties and thresholds can be leveraged to elicit a humoral response against SARS-CoV-2 via B-cell epitopes. However, it is important to note that B-cell epitope prediction is generally considered to be less reliable than its T-cell counterpart; hence the study also focused on predicting T-cell epitopes.
T-Cell epitope prediction
In the pursuit of identifying highly immunogenic peptides originating from the SARS-CoV-2 S-protein capable of eliciting CD4+ or CD8+ T-cells upon binding and presentation by MHC molecules, T-cell epitopes were predicted. It is noted that HLA variants are expressed at varying frequencies across different ethnic groups. Interestingly, diverse HLA variants can bind to similar sets of peptides and have been categorized into HLA supertypes accordingly. The correlation between HLA-A*02:01, HLA-A*02:06, HLA-A*29:02, and HLA-A*01:01 was notable in the cluster analysis of the MHC-I alleles. The IEDB recommended NetMHCpan 4.1 EL and the consensus prediction method identified MHC class-I Cytotoxic T-lymphocyte (CTL) epitopes with a size 9–10 for the HLA set including HLA class I alleles HLA-A*01:01, HLA-A*02:02, HLA-A*03:01, HLA-B*35, HLA-C*04:01, HLA-C*12:03, and HLA-DRB1*15:01. The T-cell epitopes and their corresponding amino acid positions, predicted by at least three tools, with a peptide length of 9, with a high score indicating high affinity to MHCI molecule. The top 9 epitopes were selected based on a threshold score of 0.65 from the IEDB server. Table 3 describes the selected T-cell epitopes with scores exceeding 0.65, which were subjected to further analysis. In addition, ToxinPred predicted these peptides to be non-toxin (Table 3). For more in-depth information on the IEDB NetMHCpan 4.1 EL prediction outcomes, refer to Supplementary Table 5.
An analysis utilizing a data-driven and consensus-based approach has successfully predicted nine CTL epitopes from the S-protein of SARS-COV-2. Notably, these predicted epitopes, with the exception of T-Ep7, were situated within the conserved region of the virus, a finding supported by the examination of the multiple sequence alignment (MSA) of SARS-CoV-2 variants (Supplementary Fig. 1). Employing the NetMHCII 2.3 algorithm of IEDB server revealed a total of 34 Helper T lymphocyte (HTL) epitopes specific to HLA-DQA1*01:02/DQB1*06:02. These epitopes have undergone comprehensive assessments for antigenicity, toxicity, and stability. In Tables 4 and 19 epitopes with high rank and low IC50 values are listed. Additionally, the majority of these epitopes have the potential to induce cytokines such as IL-4, IL-10, or IFN-gamma, as verified by the SVM score from the IL4pred, IL-10Pred, and IFN epitope servers. For further details, please refer to Supplementary Table 6.
Modeling and evaluation of the 3D structure of T-cell epitopes
The PEPFOLD 3.0 webserver was utilized to predict the three-dimensional structure of T-cell epitopes T-Ep1 to T-Ep5, which exhibited high CTL epitope prediction scores. Furthermore, we analyzed the Ramachandran plot (Supplementary Fig. 4) for each modeled peptide. Our observations revealed that epitopes demonstrating regular secondary structures displayed all amino acids with φ and ψ angles in the highly allowed conformation, positioned in the third quadrant. The models representing T-Ep1 to T-Ep5 were identified as conformationally stable 3D structures and were subsequently employed for binding analysis with the MHC I molecule.
Protein-protein Docking analyses suggested that the epitopes exhibit a strong binding affinity for MHC I
Molecular docking analyses were conducted using the ZDOCK tool of Discovery Studio to predict binding of five T-cell epitope peptides with MHC I molecule, PDB:7RTD. The refinement of the selected docked poses was performed using the CHARMM energy minimization algorithm of the RDOCK program to eliminate non-bond clashes and determine the best-docked pose based on RDOCK score. The binding energy (BE), ΔG, and dissociation constant (Kd) of the best-docked poses of the five epitopes were estimated. Upon detailed examinations of the docking scores and binding energy values, it was revealed that T-Ep3 and T-Ep4 exhibited the most favorable properties, demonstrating the lowest BE and Kd values for the docked complex, alongside the highest docking score and highest number of intermolecular interactions (Fig. 1; Table 5). During the docking process, T-Ep3 exhibited 20 non-bond interactions with MHC I with ZDock score and RDock scores calculated as 11.86 and − 9.99, respectively. The binding energy of the docked T-Ep3-MHC I complex was determined to be −10.3 Kcal/mol, and the dissociation constant (Kd) was found to be 2.9e-08 M. T-Ep4 also demonstrated comparable values for ZDock score (11.2), RDock score (−9.5), binding energy (−10.3 Kcal/mol), and dissociation constant (Kd = 2.7e-08). It formed 17 intermolecular contacts with the MHC I molecule (Fig. 1; Table 5). The remaining epitopes also showed significant interaction with the MHC I molecule. Moreover, it was observed that the region of T-Ep3 spanning from V687 to Y695 is situated immediately after the S1-S2 cleavage site, R685-S686 of the S protein. The SARS-CoV-2 S protein is distinct from other coronaviruses due to an insertion at the cleavage site, especially in the sequence 680SPRRAR↓SV687, resulting in a cleavage motif RxxR recognized by proteases such as furin or TMPRSS2. This efficient cleavage at the S1/S2 site is essential for the virus to enter the host target cells21,22.
Molecular docking studies using a structure-based approach revealed the efficacy of predicted T-cell epitopes in binding with MHC I molecules. Two predicted T-cell epitopes showed maximum binding capacity with the MHC I molecule, with one epitope located close to the superantigen region of the S protein of SARS-CoV-2, specifically at the site where S1-S2 cleavage occurs23. These findings emphasize the potential of the predicted T-cell epitopes in designing effective immunotherapeutic strategies.
Molecular dynamics simulation studies identified a stable conformation in the peptide-MHC I complex
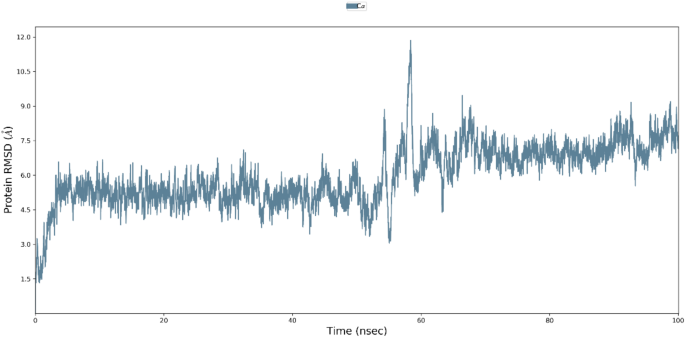
A 100 nanosecond Molecular Dynamics (MD) simulation was conducted to evaluate the stability and flexibility of the T-Ep4-MHC I complex. Trajectory files derived from the simulation were analyzed using the Root Mean Square Deviation (RMSD) to assess the deviation between all C-alpha atoms of each conformation and the reference conformation. The RMSD analysis indicated that the complex achieved a steady state within the initial 3 nanoseconds and maintained stability until 58 nanoseconds, which some minor fluctuations. The average RMSD value during this period was 5.1 Å, which subsequently increased to 7.1 Å until the completion of the 100 nanoseconds simulation (Fig. 2).
An analysis of the conformations generated of each peptide-MHC I complex revealed that the molecules remained bound throughout the simulation under physiological mimicking conditions, suggesting potential effectiveness in stimulating T-cells by sustaining MHC I binding. Examination of the docked complexes indicated no significant loss of conformation, with only minor fluctuations observed in loop regions. This discovery holds significant promise and may offer valuable insights for design and development of epitope-based vaccines.
Selection of vaccine subunits to construct a multi-epitope vaccine sequence
The vaccine subunit’s antigenic components were chosen based on their non-allergenic and non-toxic properties, their capacity to elicit immune responses, and their proximity to known epitopes.
It is noted that T-Ep1 reveals a strong MHC class I binding and CTL prediction score of 0.99, coupled with a low IC50 value of 2.78nM for the HLA-C*12:03 allele. Moreover, it demonstrates a significant binding affinity with HLA-B*35:01 and HLA-A*01:01 within the 865LTDEMIAQY873 region. Adjacent to T-Ep1, the highly ranked CTL epitope, T-Ep2 896IPFAMQMAY904, also demonstrated a score of 0.99. In the specified sequential region, an MHC class II binding HTL epitope, 886WTFGAGAALQIPFAM900, has been predicted with a favorable percentile rank of 1.5 and a lower IC50 value of 66.8nM. The close proximity of HTL epitopes suggests their potential as candidates for inclusion in a multiepitope vaccine. The proposed vaccine subunit covers T-Ep1, T-Ep2, and the HTL epitope, denoted as ‘S1’ and represented by the amino acid sequence LLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAM.
In our analysis, T-Ep4 has demonstrated significant potential, achieving a high score of 0.97 in CTL prediction and a low IC50 value of 40.59nM for HLA-C*12:03. Notably, it exhibits strong binding affinity for HLA-B*35:01 and HLA-A*01:01 in the 604TSNQVAVLY612 region. Furthermore, we have identified an HTL 602TNTSNQVAVLYQDVN616 with a commendable percentile rank (0.64) and a superior prediction score (0.20) in the same region, The amino acid sequences 604–612 display overlapping epitopes for MHC class I & II alleles, indicative of potential antigen presentation via both pathways. Our analysis also revealed a predicted linear B-cell epitope in the overlapping region 616NCTEVPVAIHADQLTPTWRVYSTGSNVFQ644. As a result of these findings, we propose these CTL, HTL, and B-cell epitopes identified as strong candidates for inclusion as multiepitope vaccine subunits. Upon integrating these findings, we suggest that the peptide 604TSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQ644, referred to as ‘S2’, has the potential to elicit T-cell and a B-cell mediated responses or both, making it a promising vaccine candidate.
The peptide, T-Ep6 440NLDSKVGGNYNYLY453 exhibited a high percentile rank in both the Artificial Neural Network (ANN) and Stabilized Matrix Method (SMM) analyses, with values of 0.05 and a low IC50 value of 22.56 nM to HLA-A*01:01 independently (Table 3). The predicted peptide for HTL response is situated in the nearby region 464FERDISTEIYQAGST478 with a percentile rank of 1.8 and prediction score of 0.12 (Table 4). Furthermore, linear and conformational B-cell epitopes, B-Ep3 have been identified within the region covering the amino acid sequence, 440NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN502 (Table 1). The B-Ep3 has a favorable PI score of 0.67 (Table 2). Of note, the Uniport database confirms the presence of an immunodominant HLA epitope region 448NYNYLYRLF456 recognized by CD8+ cells, consistent with our initial prediction. Based on these findings, it is hypothesized that the proposed peptide 440NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN502, referred to as ‘S3’ has the potential to elicit a T-cell mediated or B-cell mediated immune response, or both.
The T-Ep3, the CTL epitope anticipated within the 687VASQSIIAY695 region demonstrated a notably high prediction score of 0.97 with a percentile rank of 0.01 (Table 3). It demonstrated a strong affinity with an IC50 value of 10.63 to HLA-B*35:01 and HLA-A*01:01. Additionally, an HTL epitope, 684ARSVASQSIIAYTMS697 was identified in the same region using the consensus 2.22 IBDB prediction method (Table 4). This epitope presented a favorable percentile rank of 4.2 and an IC50 value of 135nM. Furthermore, a linear B-cell epitope 672ASYQTQTNSPRRARSVASQ690 has been forecasted in the aforementioned region (Table 1). Our study highlights the significance of T-Ep3 due to its spatial proximity to the previously recognized superantigen region, 681PRRA684of the S-protein of SARS-CoV-223. Found at the S1-S2 cleavage site, this region plays a pivotal role in initiating viral entry processes. Consequently, the peptide region 687VASQSIIAYTMS697 designated as ‘S4’ shows promise as a subunit for a promising vaccine construct.
Consequently, there are four proposed subunits for the vaccine construct, which are as follows:
S1: LLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAM
S2: TSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQ
S3: NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN
S4: VASQSIIAYTMS
Population coverage analysis
Considering the high global prevalence of SARS-CoV-2 infection, it is essential for vaccine epitopes to cover a wide range of allele populations. Our examination of population coverage for the CTL, utilizing the IEDB population coverage tool, revealed that each CTL exhibited coverage of 71%, 81%, 89%, and 90% for the Indian, Chinese, North American, and global populations, respectively, based on a specific set of HLA-A, HLA-B, and HLA-C alleles. Detailed information can be found in Supplementary Tables 7 and Table 8.
Design and characterization of multi-epitope vaccine construct
The vaccine construct was created utilizing subunits S1-S4, which were devised based on the overlapping regions within CTL, HTL, and B-cell epitopes, as well as existing literature. Human β-defensins were employed as the vaccine adjuvant at the N and C terminals, with the subunits interconnected by GPGPG linkers. Specifically, human Beta-defensin 2 (hBD-2) (PDB ID: 1FD3 sequence:
GIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKP)
and the human Beta-defensin 3 (hBD-3) (PDB ID: 1KJ6, sequence:
GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK) were utilized in this context as adjuvants at N and C terminals respectively using the linker EAAAK as illustrated in Fig. 3.
The vaccine sequence exhibits a theoretical isoelectric point (PI) of 9.19, a molecular weight of 28.2 kDa, and an estimated half-life of 30 h in vitro for mammalian reticulocytes. With an aliphatic index of 69.39, it demonstrates significant thermostability, while the instability index (II) of 32.53 categorizes the sequence as stable. Additionally, the vaccine sequence showcases a negative grand average of hydropathicity (GRAVY) of −0.22, thereby fulfilling the essential physicochemical criteria for a promising vaccine candidate.
Antigenicity, allergenicity, and toxicity of the designed vaccine construct
Upon analysis, the vaccine construct demonstrated antigenicity with a prediction score of 0.44 when assessed using the VaxiJen v2.0 tool, indicating its potential to provoke an immune response. Subsequent evaluations utilizing the AllerTop v2.0 and AllergenFPv.1.0 servers confirmed the non-allergenic properties of the vaccine construct. Furthermore, the toxin prediction tool, ToxinPred2, substantiated the non-toxic nature of the vaccine construct, yielding a machine-learning prediction score of 0.70, surpassing the established threshold of 0.60.
Cytokine-inducing ability of the MEVC
The PIP-EL server identified the construct as proinflammatory, yielding a score of 0.73. Conversely, the Pre-AIP server classified it as anti-inflammatory with a high confidence AIP score of 0.55. Moreover, the IL-10Pred server projected the construct to be IL-10-inducing with an SVM score of 0.84. Additionally, the IL4pred server indicated that nearly the entire construct region possesses IL-4-inducing ability. Also, the IFNepitope2 server predicted most of the MEVC sequence as IFN-gamma-inducing. A detailed graphical representation of IL4pred server and IFNepitope2 result is provided in Supplementary Fig. 5A-B. Given the accuracy of the three-dimensional structure in estimating vaccine construct design efficiency, we have modeled the construct for further studies.
Structure prediction and validation of designed vaccine construct
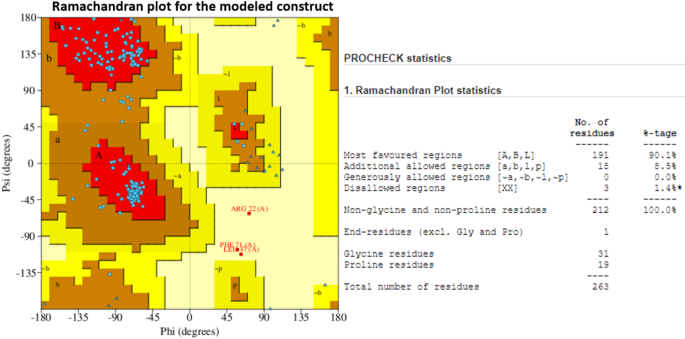
The 3-D structure of the designed construct was modeled using the trRosetta webserver. Among the refined models, the model with the highest TM score (0.335) was chosen and further refined using ModRefiner and GalaxyWeb. The final model displayed a favorable 95.4% region in the Ramachandran plot, representing the best score among all refined models (Supplementary Fig. 6). Additionally, the TM-score was measured at 0.9261, RMSD at 0.27, MolProbity at 1.8, with a minimal clash score of 11.1 and no poor rotamers (Supplementary Fig. 6). The stereochemical quality of the refined structure was validated using PROCHECK in the PDBSum. Notably in Ramachandran plot statistics, 90% of the residues were found within the ‘Most Favored regions,’ 8.5% within the ‘Additionally allowed regions,’ and only 1.4% (3 residues) fell in the ‘Disallowed regions’ (Fig. 4). These results affirm the decent quality of the model, and the conformationally stable models were employed for subsequent binding analysis with the TLR molecules24.
Molecular Docking of vaccine construct with host immune receptor
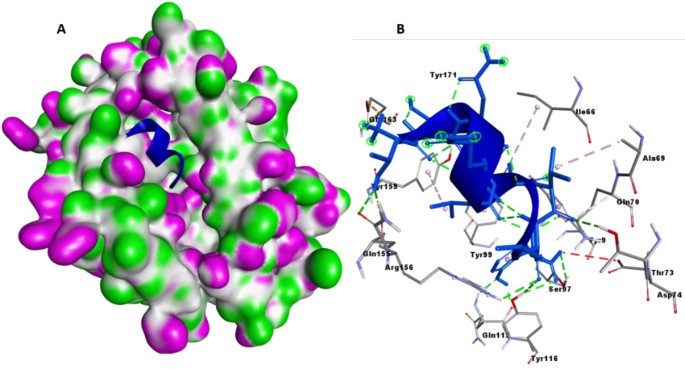
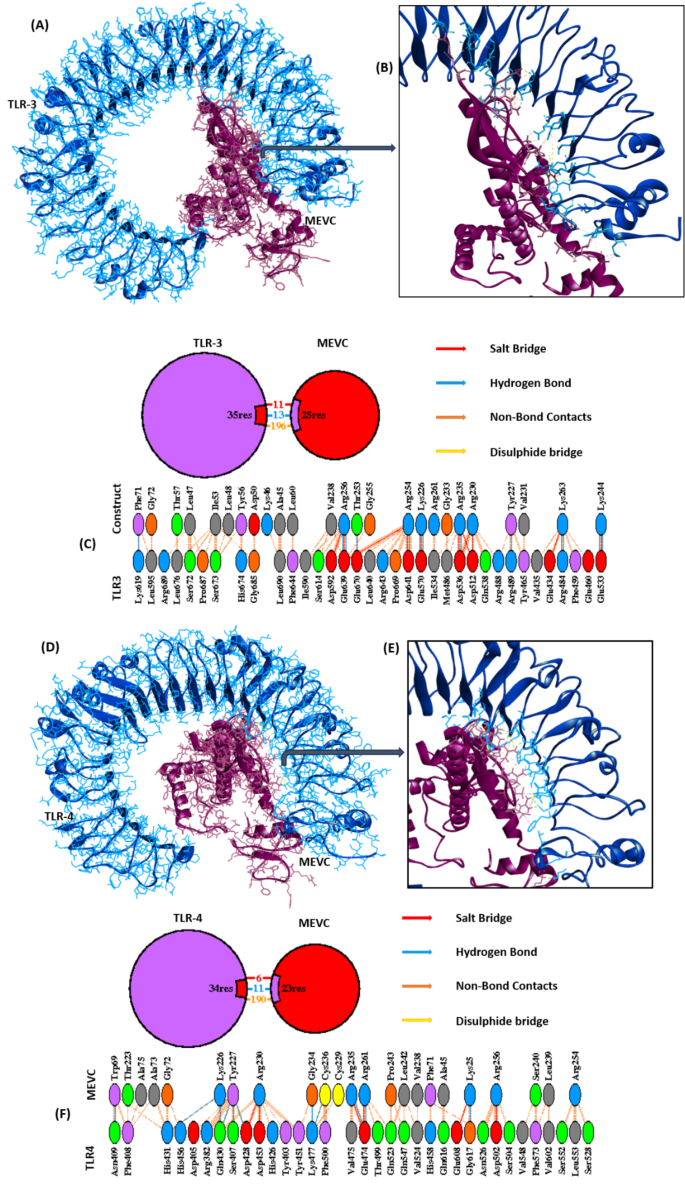
The binding efficiency of the designed construct with TLR-3 and TLR-4 was assessed using the ClusPro server. Analysis revealed that the highest number of docking poses clustered with the lowest energy-weighted score of −1303.2for TLR-3 and − 1133.6 for TLR-4, surpassing previous studies in a similar domain25. The docked complex model of MEVC-TLR-3 was examined in Discovery Studio’s molecular viewer to observe intermolecular interactions (Fig. 5A) and further analyzed in a closer view (Fig. 5B). Intermolecular interactions analysis using the Prot-Prot analysis of PDBSum estimated 11 salt bridges, 13 hydrogen bonds, and 196 non-bond contacts between MEVC and TLR-3, indicating robust binding efficiency (Fig. 5C). The same analysis was performed to examine MEVC-TLR-4 interactions, and the server computed 6 salt bridges, 11 hydrogen bonds, and 190 non-bond contacts between MEVC and TLR-4 (Fig. 5D-F). The PRODIGY server (https://bianca.science.uu.nl/prodigy/)26 predicted a Kd constant of 1.7e-12 M and 1.3e-11 for MEVC-TLR-3 and MEVC-TLR4 complex, respectively. The binding free energy (ΔG) of −16.0 Kcal/mol and − 14.8 Kcal/mol were computed for the MEVC-TLR-3 and MEVC-TLR4 complexes, respectively, highlighting the strong binding between the two molecules in both complexes. We also performed docking of MEVC with TLR4-dimer, and the docked complex (lowest energy-weighted score of −1060.6) was bound with an estimated Kd value of 2.3e-13 M and BE value of −17.3Kcal/mol. Detailed examination showed that MEVC formed many contacts with TLR-4 dimer Chain A (1 salt bridge, 2 hydrogen bonds, and 64 non-bonded) and Chain B (6 salt bridges, 13 hydrogen bonds, and 148 non-bonded contacts), indicating strong binding efficiency of MEVC with TLR-4 also. Results are provided as supplementary details (Supplementary Fig. 7). The calculated binding energy value, docking score, and the number of intermolecular contacts collectively suggest that the engineered peptide construct, MEVC, exhibits a strong binding affinity with the TLR-3 and TLR-4 molecules. This finding underscores the efficacy of the construct in its intended function.
The docked complex model of the MEVC-TLR-3 and MEVC-TLR-4. (A and D) The docked complex MEVC-TLR-3 and MEVC-TLR-4-model as examined in the molecular viewer of Discovery Studio, (B and E) A closer view of the MEVC-TLR-3and MEVC-TLR-4complex, (C and F) Intermolecular interactions between MEVC and TLR-3 and MEVC and TLR-4 in the docked complex structure of Prot-Prot Analysis.
Dynamic simulation of MEVC-TLR-3 complex
Dynamic simulation study of the MEVC-TLR-3 complex was performed using the iMODS online server to assess and measure the proteins’ flexibility. From the analysis reports provided as supplementary information (Supplementary Fig. 8), the deformability peaks in the graph of the vaccine-TLR3 complex indicate flexible regions in the protein (Supplementary Fig. 8a). The eigenvalue of the vaccine-receptor complex is 5.399225e-06, representing the protein’s stiffness (Supplementary Fig. 8b). Variance and covariance graphs show associated amino acids and rigid regions in the complex (Supplementary Fig. 8c). The elastic map illustrates the connections between atoms and highlights rigid regions in the complex (Supplementary Fig. 8d).
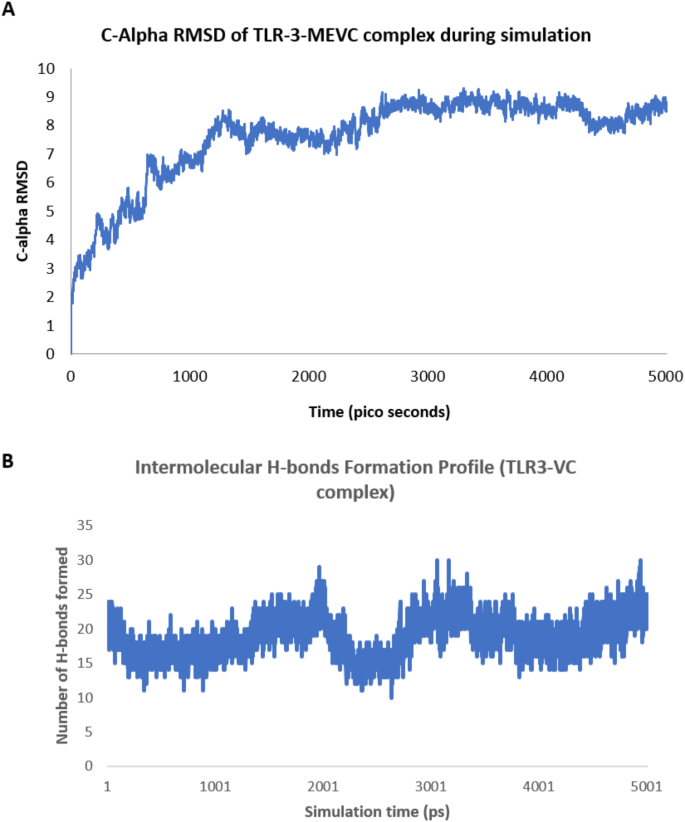
An all-atom Molecular Dynamics (MD) simulation was conducted for 50 ns to evaluate the stability and flexibility of the MEVC-TLR-3 complex. Trajectory files derived from the simulation were analyzed for the Root Mean Square Deviation (RMSD) to assess the deviation between all C-alpha atoms of each conformation and reference starting conformation. The C-alpha RMSD was computed for MEVC-TLR-3complex to examine the deviation in conformation during the simulation period. The RMSD analysis indicated that the complex gradually achieved a steady state trend in the initial 15 nanoseconds and maintained a stabilized nature until the 50 nanoseconds, with very minor fluctuations. The average RMSD value during this period was 7.5 Å, subsequently increasing to 8.5 Å until the completion of the 50 ns simulation (Fig. 6A), which indicates that the binding of MEVC-TLR-3 is strong enough and the complex remains in the stable bound state. The number of hydrogen bonds formed between TLR-3 and MEVC is computed throughout the simulation period and plotted (Fig. 6B). It was found that an average of 20 intermolecular H bonds were formed in the MEVC-TLR3 complex which indicates how strongly the molecules are bound to each other throughout the simulation period. The binding free energy (ΔG) for the last generated MEVC-TLR-3 conformation was computed as −15.8 Kcal/mol and the dissociation constant, Kd as 2.6 e-12 M which is almost the same as that of the initial conformation (BE= −16.0 Kcal/mol and Kd = 1.7e-12 M) indicating that molecules remained strongly bound even after the 50 nano seconds of simulation time. Molecular Dynamics (MD) simulation of MEVC-TLR-3 complex revealed that the molecules remained strongly bound during the simulation, indicating the potential effectiveness of our vaccine construct in TLR-3-mediated immune response.
C-alpha RMSD plot and Intermolecular H-bond profile of MEVC-TLR3 complex. (A) C-alpha RMSD plot for 50 ns simulation period to analyze the stability of the MEVC-TLR-3 complex. The RMSD values, in Å, of the C-atoms of each MD-generated conformation in comparison to the reference conformation is plotted over50 ns. (B) Hydrogen bond formation between the TLR-3 and MEVC during the simulation period.
Codon adaptation and in Silico cloning of the vaccine construct
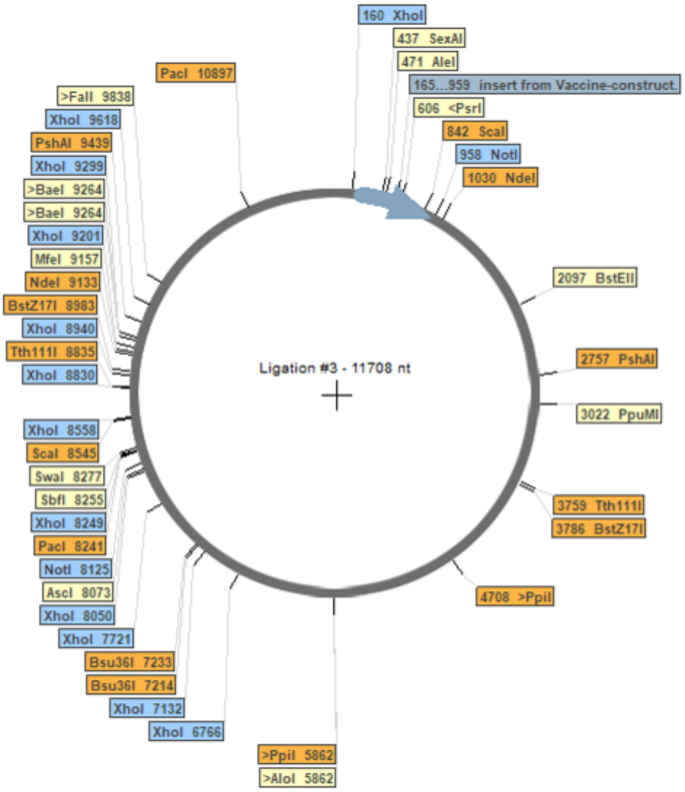
The Java Codon Adaptation Tool was employed to optimize the codon usage of the vaccine, resulting in a 789-nucleotide sequence with a Codon Adaptation Index (CAI) of 1.0 and a GC content of 53.73%. These findings suggest the potential for expression of the vaccine construct in the host cell, E. coli. The CAI value indicates the likelihood of cDNA expression in a specific expression system. To facilitate cloning, XhoI and NotI restriction sequences were incorporated at the N and C termini of the optimized codon sequence. Furthermore, the optimized sequence was integrated into the pET-28(+) vector using Serial Cloner 2.6 software for in silico cloning (Fig. 7). This operation successfully inserted the optimized codon sequence of the vaccine construct into the plasmid following cleavage at the restriction enzyme sites (Fig. 7).
In Silico immune simulation studies in response to our vaccine injection
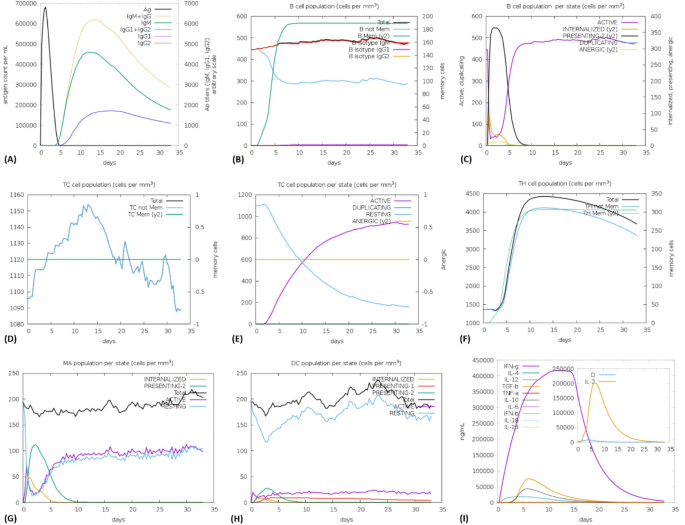
The designed vaccine construct was subjected to in silico testing to assess its impact on the immune system’s response. The simulation conducted using the C-IMMSIM server indicated that upon administration of the vaccine construct, the initial response showed elevated levels of IgM + IgG. Subsequently, the secondary response displayed increased levels of IgM, IgG1, and B-cell populations within the 10–15-day period (Fig. 8A-C). It is noteworthy that the antigen level (680000 counts per mL) decreased significantly by the 3rd day following the injection. Moreover, the B-cell response demonstrated a logarithmic growth phase from day 3 to day 7, reaching a peak level of approximately 600 cells per mm3. This underscores the vaccine construct’s ability to stimulate substantial levels of B cells that produce IgM and IgG subclass antibodies. Both CTL and HTL elicited a robust immune response after a brief exposure, indicating the immunogenic nature of the T cell epitopes integrated into the vaccine construct (Fig. 8D-F). Additionally, the populations of macrophages and dendritic cells were observed to increase post-exposure (Fig. 8G-H). Our research also revealed a sharp increase in the production of pro-inflammatory cytokines such as IFN-γ, IL-2, and IL-12 post-vaccination (Fig. 8I). Concurrently, peaks in certain anti-inflammatory cytokines such as TGF-β and IL-10 were noted, indicating a reliable and crucial immune-triggering response upon the administration of the vaccine construct (Fig. 8H-I).
In silico simulation of immune response after the administration of the vaccine as an antigen. (A) levels of antigen and immunoglobins. (B) density of B-cell population. (C) density of B-cell population per state. (D) density of cytotoxic T-cell population. (E) density of cytotoxic T-cell population per state. (F) density of helper T-cell population. (G) density of macrophages population per state. (H) density of dendritic cell population per state. (I) level of cytokine production.