Platform overview
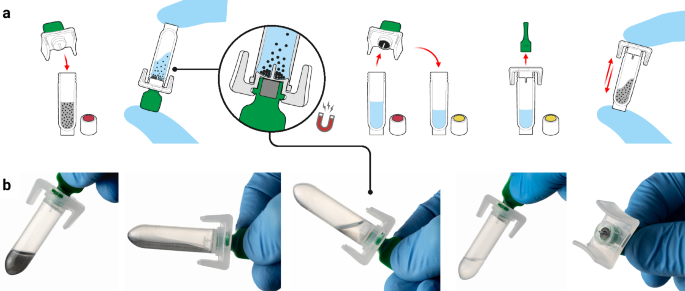
The developed platform is comprised of three core components: a single-use sample extraction kit, a lyophilised colourimetric LAMP-based panel, and a low-cost isothermal heat block, as shown in Fig. 1a. To support a POC workflow, the platform also includes a sample collection kit (containing a swab and inactivating medium, COPAN eNAT®), a reusable fixed-volume pipette with disposable tips, and an optional tablet with cloud-connected companion software for result logging and centralised data management. The Dragonfly workflow includes power-free extraction of nucleic acids in under 5 minutes, followed by colour-based molecular detection in less than 35 minutes, culminating in a visual equipment-free result read-out. A high-level overview of the complete workflow is shown in Fig. 1b, c, with the core nucleic acid extraction technology, SmartLid, highlighted in Fig. 2, and photographs of select process steps, from sample input to result readout, provided in Fig. 3a–f.
a Overview of the platform, including consumable components, optional tablet and companion app, as well as deployed disposable mobile workstation, and low-cost isothermal heat block with reusable fixed-volume pipette. b High-level overview of rapid nucleic acid extraction process and test panel loading for one sample. c High-level overview of test panel incubation and colourimetric result interpretation. Created in BioRender. Cavuto, M. (2025) https://BioRender.com/d45n457.
a Graphical illustration of SmartLid usage to transfer magnetic beads from one tube to another with a removable magnet. b Close-up images showing the rapid magnetic bead collection process, with beads visible on the underside of the SmartLid after collection. Created in BioRender. Cavuto, M. (2025) https://BioRender.com/d45n457.
a Sample input using a disposable exact-volume pipette. b Insertion of the SmartLid to initiate the rapid power-free extraction process. c Close-up of extracted nucleic acids being loaded into the open test panel using a 20 µL fixed-volume pipette. d Simultaneous incubation of two test panels, with a capacity of up to four panels per low-cost, portable isothermal heat block. e Fully incubated colourimetric test panel being removed from the heat block. The attached panel tag allows for easy removal, reduces the risk of accidental tube opening, and verifies correct panel selection through a comparison with the scanned tag at the beginning of the process. f Example of a test panel result indicating a positive (yellow) reaction for OPXV, along with valid test controls. Screenshots from the companion application, showing the test panel loading screen (g), incubated test panel result capture screen (h), result confirmation screen (i), and result interpretation and recording screen (j). Created in BioRender. Cavuto, M. (2025) https://BioRender.com/d45n457.
Platform components
Previously developed by our group48,49 and as depicted in Fig. 2, the SmartLid technology utilises a magnetic lid to capture and transfer superparamagnetic nanoparticles (or magnetic beads) and attached DNA/RNA through three simple steps (lysis-binding, washing, and elution), enabling efficient and power-free nucleic acid extraction without centrifugation or manual pipetting. All required buffers are pre-aliquoted in colour-coded tubes (utilising a sequential traffic-light system of red, yellow, and green) and packaged in a cardboard tray (150 × 70 × 50 mm) along with disposable exact-volume pipettes for sample input and a SmartLid for magnetic bead manipulation. In Fig. 3c, we show that the fully recyclable carboard packaging also functions as a workstation, facilitating the POC workflow with clear diagrammatic labelling, a space for housing the sample tube, and receptacles for holding the lyophilised test panel during reagent resuspension.
For the test panel, a lyophilised colourimetric LAMP chemistry was developed to yield long term room temperature storage, visual result readout, and isothermal incubation50. This enables visual identification of positive reactions based on proton production (and subsequent pH drop), which occurs with nucleotide incorporation during DNA polymerase activity. Furthermore, this colour transition, from pink (at high pH) to yellow (at low pH), is compatible with multiple colour vision deficiency (CVD) friendly colour schemes, ensuring maximum usability regardless of user profile51. An off-the-shelf eight-tube PCR strip (4titude®), with individually flip-capped 0.2 mL tubes, was selected to house each pre-dispensed and lyophilised reaction master mix. To enhance traceability, polycarbonate tags, attached in the middle of each tube strip, provide space for data-matrix and alpha-numeric labelling. Combined, these three aspects create a platform that minimises reliance on equipment, requiring only a low-cost, portable, and user-friendly isothermal heat block (160 × 110 × 130 mm, <1 kg). This heat block can be powered by mains electricity, a standard 12-volt supply, batteries, or solar panels, drawing less than 20 W continuously once at the correct temperature.
Cloud-connected companion application
To augment the manual workflow, an optional companion Android application was developed with the goal of improving the user experience and enabling wireless cloud connectivity to integrate with healthcare databases (Fig. 3g–j). Utilising the onboard device camera, the application further enhances traceability by scanning data matrices on all consumables at the start of the process. A virtual step-by-step workflow was implemented to guide new users through the process, with progress tracked and displayed, and animated clickable timers provided throughout the workflow to time each step (e.g., buffer shaking). Test panel incubation is also monitored and timed, with assistance provided to remind the user of the incubation status of each sample and their respective location in the heat block. After incubation, result interpretation and recording were augmented through an image capture process, in which a cropped and enlarged view of the eight tubes is presented along with buttons to select observed colours. Depending on the specific panel scanned in at the beginning of the process, results are then interpreted and displayed. All results, along with captured test panel images, are automatically stored and summarised in an AWS cloud-accessible dashboard for export, review, and traceability. Further details of the Dragonfly application are provided in Supplementary Methods.
Test Panel Design
Our Skin Infection Viral Test Panel was designed to target orthopoxvirus genus (OPXV), monkeypox virus (MPXV), varicella-zoster virus (VZV), herpes simplex virus type 1 (HSV-1), and herpes simplex virus type 2 (HSV-2). VZV, HSV-1 and HSV-2 can cause skin rashes and lesions potentially confounding mpox diagnosis. In addition, zoonotic OPXVs such as cowpox and borealpox have been associated with fatal human infections in Europe and America52. Therefore, our panel aims to facilitate the accurate diagnosis of mpox cases while capturing the emergence of zoonotic OPXV species. The panel layout is shown in Fig. 1c and includes the following targets, from left to right: colour reference control, OPXV, MPXV, VZV, HSV-1, HSV-2, extraction control, and internal control.
Two LAMP assays targeting two distinct genomic regions were included per lyophilised reaction mix for OPXV, MPXV, and VZV, with one assay each for HSV-1 and HSV-2. For detection of OPXV, we selected E9L (viral DNA polymerase gene, a conserved segment across all Eurasian OPXVs) and F13L (encoding a conserved protein essential for viral maturation and release from infected cells53), with E9L also used as the target in the US Centers for Disease Control and Prevention (CDC) qPCR assay54. For MPXV, assays targeting conserved intraspecies regions of G2R (a viral tumour necrosis factor receptor) and A9L (a VACV orthologue encoding a morphogenesis factor) were designed to cover both clade I and II without cross-reactivity with other OPXVs. G2R is the target currently used by the CDC qPCR assay for MPXV detection55. For VZV, an assay targeting the ORF28 gene was designed, along with an additional assay targeting the ORF62 gene, as described by Okamoto et al.56. For HSV-1, an assay from Kaneki et al.57 targeting the UL1 gene was used, and for HSV-2, we developed an assay targeting the US4 gene. LAMP assay sequences are provided in Supplementary Table 2.
The panel includes three additional control reactions to ensure reliable results. First, as Dragonfly utilises a pH-based colourimetric indicator, variations in sample and extracted elution pH were observed to affect the starting resuspended (negative) reaction colour. To account for this, a “colour reference control” was added (which excludes amplification enzymes), ensuring that one tube always remains pink, and thus represents a pH-adjusted negative reference colour. Next, an “internal control” reaction mix was added, which contains the DNA template specific to its assay, ensuring amplification under ideal operating conditions. This control acts as a confirmatory reaction to indicate that the test panel is in good working order (e.g., not damaged due to improper storage, incubated at the wrong temperature, or excessively inhibited). Finally, a “human extraction control” was included, targeting the human housekeeping beta-actin gene58, providing a confirmation that the extraction process was performed correctly and that the sample source was adequately swabbed. A valid test result, whether positive or negative, requires that the three described control reactions are pink (negative), yellow (positive), and yellow (positive), respectively.
Workflow
The nucleic acid extraction and molecular detection protocols were optimised to achieve a balance between workflow complexity, time, and performance: Inactivated swab eluent (400 µL) is transferred from the sample collection tube into Tube A (containing lysis-binding buffer and magnetic beads) using a disposable exact volume pipette (Fig. 3a). The included SmartLid is then used to transfer magnetic beads and their attached nucleic acids through a series of three sample extraction steps (A: lysis-binding, B: wash, and C: elution) (Fig. 3b). At each step, resuspended magnetic beads are mixed with the buffers for 30 seconds through manual shaking without the magnet, followed by insertion of the magnet to collect the magnetic beads, and a 30 second drying step to evaporate any remaining solvents prior to elution. Once nucleic acids are eluted into Tube C, the magnetic beads are removed with the SmartLid. Next, a 20 µL fixed volume pipette (using a disposable tip) is used to transfer elution into each tube of the test panel, resuspending the lyophilised reagents (Fig. 3c). Unlike laboratory micro-pipettes, the included fixed-volume pipettes were modified to remove the secondary ‘blow-out’ stage, simplifying the pipetting process without affecting test sensitivity or repeatability. Once all caps are firmly closed, test panels are placed into one of the four heat block rows (Fig. 3d) and incubated for 35 minutes at 63.5 °C (heater locked at temperature to reduce potential user error). After incubation, test panels are placed into a result capture card, where developed colours can be compared to a key directly below each tube, indicating the result: pink for negative, yellow for positive. (Fig. 3e, f) The entire process, from sample-to-result, was optimised to take <40 minutes per sample, with each subsequent sample able to be extracted while previous samples are being incubated, yielding a continuous throughput of greater than 12 samples per hour per user.
Analytical sensitivity and specificity of LAMP assays
Analytical sensitivity was evaluated using serial dilutions of synthetic DNA (one for each target), and the obtained standard curves had a correlation (R2) of 80 to 99% (shown in Supplementary Fig. 1). Assay optimisation results are provided in Supplementary Data, assays sequences in Supplementary Table 2, and synthetic DNA sequences in Supplementary Table 4. Time-to-positive (TTP) values across all tested concentrations ranging from 101 to 107 copies per reaction were within 15 minutes except for HSV-2, which required 25 min. All assays had an overall limit of detection (LOD) from 10 to 500 copies per reaction as shown in Fig. 4b and Supplementary Table 3. Analytical specificity was experimentally assessed using extracted nucleic acids from commercially available viral particles (Vircell, MBTC032-R, Zeptometrix CATALOG# NATHSV-6L and Zeptometrix CATALOG# NATVZV-STQ). As shown in Fig. 4c, the LAMP assays only amplified their specific targets.
a Evaluation of virucidal activity of eNAT® against vaccinia virus (VACV): Plaque assays were performed using confluent monolayers of BSC40 cells. The bar plot shows the virucidal activity after a 2-minute exposure, showing 8-log reduction, alongside images of crystal violet-stained plaque assays (1% crystal violet, 70% ethanol) after 30 minutes. b Analytical sensitivity of LAMP assays: range of detected concentrations of synthetic DNA (log10 of copies per reaction) and corresponding TTP values in minutes. c Analytical specificity of LAMP assays: Using extracted nucleic acids from various commercially available viral particles (MXPV, HSV-1/HSV-2, and VZV), and a control sample consisting of synthetic DNA at 5 × 10³ copies per reaction. The bar plot displays mean TTP values and data points (in minutes) with error bars representing the standard deviation (SD). d qPCR Ct values distribution: A histogram illustrating the distribution of qPCR Ct values obtained from OPXV and MPXV-positive clinical samples (purple dots for OPXV and yellow dashed lines for MPXV). Confusion matrices of the diagnostic performance are included below. e Sample-to-result demonstration: Images showing the results for vaccinia (VACV), cowpox (CPXV), MPXV, and combined HSV-1/HSV-2/VZV viral particles, with spiking concentrations indicated below each set of reactions. VACV and CPXV viral particles, both spiked at 5 × 105 PFU/mL, demonstrate the specificity of the OPXV assay, showing no cross-reactivity with other target assays. Similarly, spiking MPXV (2.5 × 10³ copies/mL) and combined HSV-1/HSV-2/VZV viral particles (2.5 × 10³ copies/mL for VZV and HSV-1, 3.5 × 10³ copies/mL for HSV-2) showed no cross-reactivity. Created in BioRender. Cavuto, M. (2025) https://BioRender.com/d45n457.
Sample-to-result evaluation with viral particles
Commercially available MPXV and HSV-1/HSV-2/VZV viral particles (Vircell, MBTC032-R and MBTC016) were spiked into eNAT® (COPAN) inactivation buffer at various concentrations, as shown in Supplementary Table 5. Extractions were performed using Dragonfly Sample Preparation Kits, with eluted nucleic acids used to resuspend our Skin Infection Viral Test Panels, which were then incubated for 35 minutes. The LOD for OPXV and VZV was determined to be 50 copies per reaction, equivalent to 1.25 × 103 copies per mL, assuming 100% extraction efficiency. The LODs for MPXV and HSV-1 were determined to be 100 copies per reaction, and 143 copies per reaction for HSV-2. Assuming 100% extraction efficiency, this corresponds to 2.50 × 103 and 3.58 × 103 copies per mL for MPXV/HSV-1 and HSV-2, respectively. Examples of these results are shown in Fig. 4e. Lastly, specificity of the panel was further confirmed using cowpox (CPXV) and vaccinia (VACV) viral particles at concentrations of 5×105 PFU per mL, where appropriately only the OPXV reactions turned yellow.
Virucidal activity of eNAT® buffer against VACV and HSV-1
As shown in Fig. 4a, after 2 minutes of incubation at room temperature, eNAT® buffer demonstrated significant virucidal activity against VACV, achieving a reduction in viral titters by ≥8.0 log10 TCID50/mL. This rapid inactivation indicates that the buffer is highly effective at neutralising the virus under the tested conditions, suggesting it is suitable for safe sample handling and processing. Additionally, the eNAT® buffer exhibited potent virucidal activity against HSV-1. Following a slightly longer incubation period of 5 minutes at room temperature, viral titters were reduced by ≥7 log10 TCID50/mL, further confirming the buffer’s efficacy across multiple virus families. See Supplementary Fig. 2 for a summary of the results.
Testing of clinical skin lesion swabs
A total of 164 surplus extracts from skin lesion swabs, which had been submitted to North West London Pathology (NWLP) for clinical testing, were utilised to assess whether the described viral DNA targets could be detected using the Dragonfly platform. These results were compared with identifications obtained via gold-standard automated nucleic acid extraction and real-time multiplex qPCR detection. The samples, collected in Roche COBAS PCR media (P/N: 07958030190) included 51 mpox clade II positive samples and 40 samples positive for one or more herpes simplex virus. Our platform demonstrated high analytical performance, with 96.1% (95% CI of 86.5% to 99.5%) sensitivity and 100% (95%CI of 96.8% to 100%) specificity for OPXV, and 94.1% (95%CI of 83.8% to 98.8%) sensitivity and 100% (95%CI of 96.8% to 100%) specificity for MPXV. Distributions of the Ct values for positive samples are shown in Fig. 4d. There were two false negatives for OPXV and MPXV, both of which had qPCR Ct values above 33 (34.37/33.61 and 35.96/34.97, for OPXV and MPXV respectively) and one false negative exclusively for MPXV with a qPCR Ct value of 28.65. The complete dataset, with results for all targets and clinical samples, is provided as Supplementary Data.
The swabs included 10 VZV, 20 HSV-1, and 10 HSV-2 infections, as identified by qPCR, representing 24.4% (40/164) of the samples analysed. This included 4/164 (6.3%) co-infections with mpox (three with HSV-1, and one with HSV-2). The Dragonfly platform detected 9/10 VZV, 18/20 HSV-1, and 7/10 HSV-2 cases (85% of the positive samples), including three out of the four co-infections. Confusion matrices of diagnostic performance are provided in Supplementary Fig. 3. All samples yielded a positive result for the human extraction control reaction (targeting the beta-actin gene), confirming the quality of the extraction and the adequacy of sample collection, limiting the potential for false negatives due to user error or insufficient sampling. To verify the results of the human extraction control reaction, a TaqMan assay targeting the RNase P gene was used for the qPCR comparator, with a distribution of Ct values shown in Supplementary Fig. 4.