Viral historical past unlocked: How historic museum specimens are rewriting the story of DNA virus evolution and their connections to immediately’s outbreaks.
Research: Screening great ape museum specimens for DNA viruses. Picture Credit score: Jan Krava / Shutterstock
Scientists on the College of Vienna, Austria, have screened nice ape specimens obtained from pure historical past museums to determine DNA viruses. This groundbreaking research offers distinctive insights into the historic viromes of nice apes and their potential hyperlinks to fashionable viral lineages. The findings might be of worth for learning DNA virus evolution.
The research is revealed within the journal Scientific Reports.
Background
Pure historical past museums protect an unlimited assortment of fossils and specimens, which function helpful scientific assets for learning genetic range, ecosystem dynamics, and evolutionary processes.
Museomics is the research of historic or historic DNA specimens preserved in museums. This revolutionary discipline leverages fashionable DNA sequencing methods to reconstruct historic and historic genomes, providing a deeper understanding of biodiversity by means of time. These specimens can be utilized to reconstruct historic genomes utilizing high-throughput DNA sequencing methods.
The research of infectious ailments is an attention-grabbing space in Museomics. Historical microbial genetic supplies have immense potential to make clear the evolution and transmission of infectious pathogens.
Viral genomes reconstructed from archaeological specimens have proven that nonhuman primates, resembling nice apes, host a broad vary of viruses with a DNA genome (DNA viruses). Co-phylogenetic analyses have proven that DNA viruses are stably related to their nice ape hosts for an intensive time interval. These findings underscore the evolutionary interactions between host species and their related viruses.
On this research, scientists screened 209 nice ape museum specimens utilizing high-throughput DNA sequencing to determine DNA viruses. They included each wild and captive nice ape species of their evaluation.
Particularly, they developed and applied a bait set that lined a complete of 99 totally different DNA viruses belonging to 13 viral households.
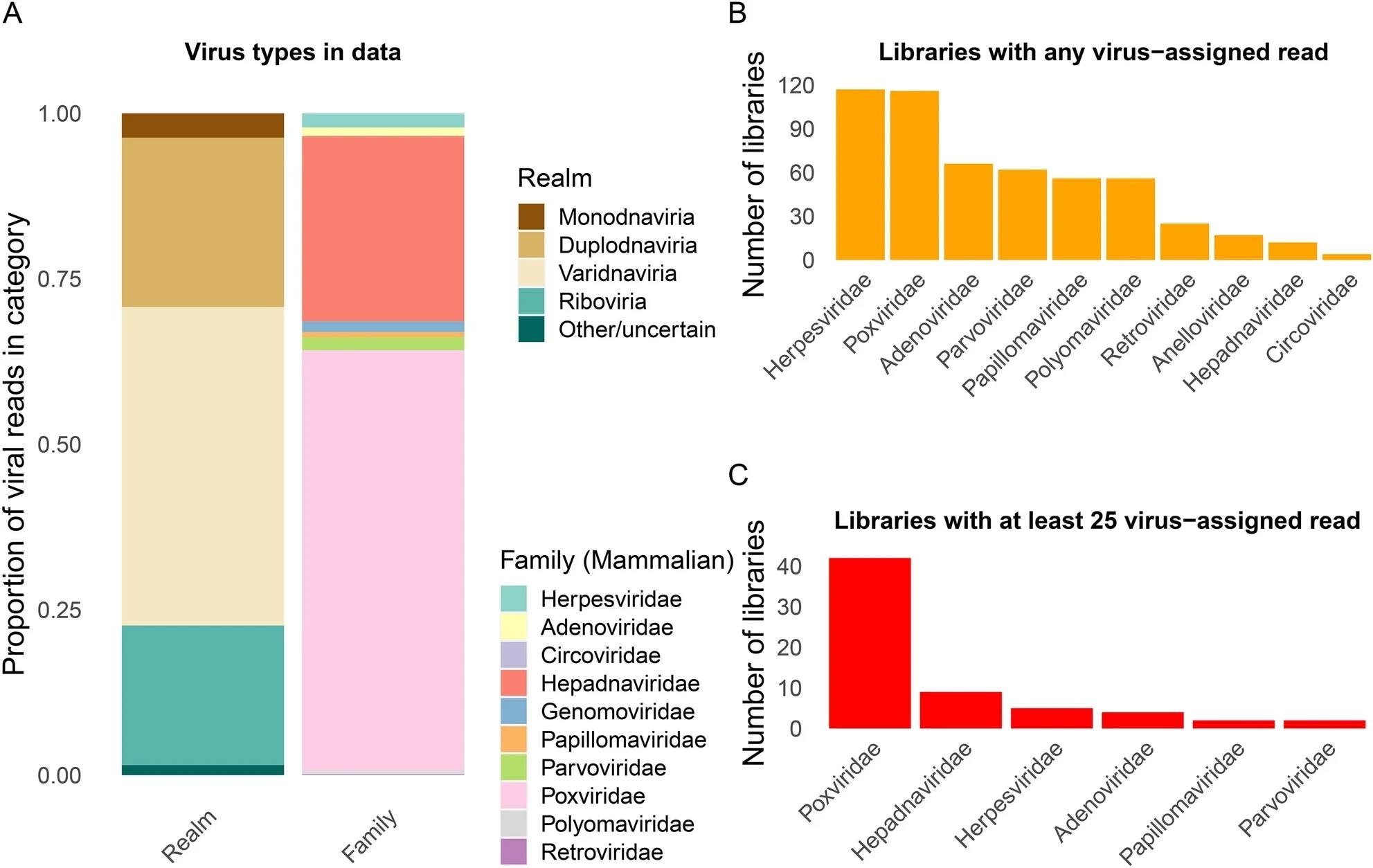
Summaries of reads assigned to totally different virus domains and households. (A) Proportion of virus-assigned reads primarily based on kraken2 throughout 214 libraries, stratified by realm and by household (for DNA viruses and Retroviridae). (B) Variety of libraries with any learn assigned to virus households. (C) Variety of libraries with at the least 25 reads assigned to virus households.
Essential observations
Scientists extracted DNA from nice ape museum specimens and sequenced it in quest of DNA viruses. The extracted DNA was much less fragmented than historic DNA, as historic museum specimens are usually youthful than archaeological specimens.
Excessive-throughput DNA sequencing detected the presence of a number of viruses from museum specimens. Six full viral genomes have been obtained, together with three hepatitis B virus genomes and three monkeypox virus genomes. Hepatitis B virus genomes have been recognized in a single gorilla and two chimpanzees, and monkeypox virus genomes have been recognized in three orangutans.
The high-coverage hepatitis B virus genome was similar to the genome sequenced beforehand from the blood pattern of a wild-born western lowland gorilla from southern Cameroon. The gorilla specimen analyzed within the present research was additionally from Cameroon, sampled greater than 40 years in the past. This continuity of viral lineage demonstrates the steady affiliation between sure viruses and their host species over time.
The opposite two hepatitis B viral genomes obtained from chimpanzees exhibited a deeper divergence from the closest recognized reference genomes.
Two hepatitis B genomes sequenced on this research, one from a wild-born Nigeria-Cameroon chimpanzee, have been similar to the genome beforehand sequenced from the specimen of a chimpanzee from Angola, a Southern African nation.
One other hepatitis B genome from a chimpanzee from central Cameroon confirmed the closest match to the genome reported from Conkouati-Douli Nationwide Park, Congo.
Three orangutans yielding monkeypox virus genomes have been from a zoo in Sumatra, an Indonesian island. Nevertheless, the zoo’s identify was not specified within the museum paperwork. Scientists said that there was a monkeypox outbreak in Rotterdam Zoo in 1964–1965, throughout which six out of 9 contaminated orangutans died.
The genetic similarity of the monkeypox genomes sequenced on this research to these from the Rotterdam outbreak strongly means that the sampled orangutans have been victims of this epidemic.
Research significance
The research highlights the presence of a number of DNA viruses in specimens of nice apes obtained from pure historical past museum collections.
Essentially the most notable discovering is the detection of high-coverage (better than 18-fold) hepatitis B virus genomes from one gorilla and two chimpanzees. The reconstruction of near-complete viral genomes from these specimens and their placement within the phylogenetic context offers proof that these viral strains are certainly related to their respective host species and associated to presently circulating viral lineages.
Total, the research demonstrates the probability of viral DNA enrichment and detection from museum specimens of nice apes. A research of this type might present info on the charges of an infection or patterns of transmission between people and different species.
Scientists confronted challenges resembling the supply of viral strains inside databases and using goal enrichment, which restricted the chance of detecting undescribed historic strains. One other concern was the absence of focused viruses in some libraries.
However, the research showcases the effectiveness of goal enrichment methods, reaching greater than a 200-fold enrichment of viral DNA. Nevertheless, the goal enrichment technique used within the research is environment friendly and cost-effective in detecting and reconstructing previous viral unsampled range, given enrichment elements of greater than 200-fold.
Scientists spotlight the necessity for spatiotemporal research of the historic virome utilizing massive datasets of viral genomes. Such research might illuminate patterns of viral transmission and host interplay throughout geographic and temporal scales, aiding in understanding zoonotic dangers and conservation efforts.
Journal reference:
- Hämmerle, M., Guellil, M., Cheronet, O., Sawyer, S., Lizano, E., Rymbekova, A., Gelabert, P., Bernardi, P., Han, S., Rattei, T., Schuenemann, V. J., Guschanski, Okay., Pinhasi, R., & Kuhlwilm, M. (2024). Screening nice ape museum specimens for DNA viruses. Scientific Experiences, 14(1), 1-10. DOI: 10.1038/s41598-024-80780-w, https://www.nature.com/articles/s41598-024-80780-w
![[original_title]](https://rawnews.com/wp-content/uploads/2024/12/ImageForNews_797145_17331900514207788-1024x682.jpg)